200 Genetic Disequilibrium
Genetic Linkage and Distances
Linked genes can become unlinked during recombination; the probability of genes separating depends on their distance from each other.
Learning Objectives
Discuss how linked genes can be inherited separately
Key Takeaways
Key Points
- Alleles positioned on the same chromosome are not always inherited together because during meiosis linked genes can became unlinked.
- Frans Janssen suggested chromosomes become unlinked during homologous recombination, a process where homologous chromosomes exchange segments of DNA.
- Alfred Sturtevant hypothesized that alleles that were closer together on a gene were more likely to be inherited together rather than alleles that were farther apart and used measurements of recombination between genes to create the first genetic map.
- When genes are perfectly linked, they have a recombination frequency of 0.
- When genes are unlinked, they have a recombination frequency of 0.5, which means 50 percent of offspring are recombinants and the other 50 percent are parental types.
Key Terms
- homologous recombination: a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of DNA
- linkage: the property of genes of being inherited together
- synapsis: the association of homologous maternal and paternal chromosomes during the initial part of meiosis
Genetic Linkage and Distances
Mendel’s work suggested that traits are inherited independently of each other. Morgan identified a 1:1 ratio between a segregating trait and the X chromosome, suggesting that the random segregation of chromosomes was the physical basis of Mendel’s model. This also demonstrated that linked genes disrupt Mendel’s predicted outcomes. The fact that each chromosome can carry many linked genes explains how individuals can have many more traits than they have chromosomes. However, observations by researchers in Morgan’s laboratory suggested that alleles positioned on the same chromosome were not always inherited together. During meiosis, linked genes somehow became unlinked.
Homologous Recombination
In 1909, Frans Janssen observed chiasmata (the point at which chromatids are in contact with each other and may exchange segments) prior to the first division of meiosis. He suggested that alleles become unlinked when chromosomes physically exchange segments. As chromosomes condensed and paired with their homologs, they appeared to interact at distinct points. Janssen suggested that these points corresponded to regions in which chromosome segments were exchanged. It is now known that the pairing and interaction between homologous chromosomes, known as synapsis, does more than simply organize the homologs for migration to separate daughter cells. When synapsed, homologous chromosomes undergo reciprocal physical exchanges of DNA at their arms in a process called homologous recombination, or more simply,”crossing over.”
Genetic Maps
In 1913, Alfred Sturtevant, a student in Morgan’s laboratory, created the first “chromosome map,” a linear representation of gene order and relative distance on a chromosome.To construct a chromosome map, Sturtevant assumed that genes were ordered serially on threadlike chromosomes. He also assumed that the incidence of recombination between two homologous chromosomes could occur with equal likelihood anywhere along the length of the chromosome. Operating under these assumptions, Sturtevant hypothesized alleles that were far apart on a chromosome were more likely to dissociate during meiosis simply because there was a larger region over which recombination could occur. Conversely, alleles that were close to each other on the chromosome were likely to be inherited together. The average number of crossovers between two alleles, or their recombination frequency, correlated with their genetic distance from each other, relative to the locations of other genes on that chromosome. Sturtevant divided his genetic map into map units, or centimorgans (cM), in which a recombination frequency of 0.01 corresponds to 1 cM.
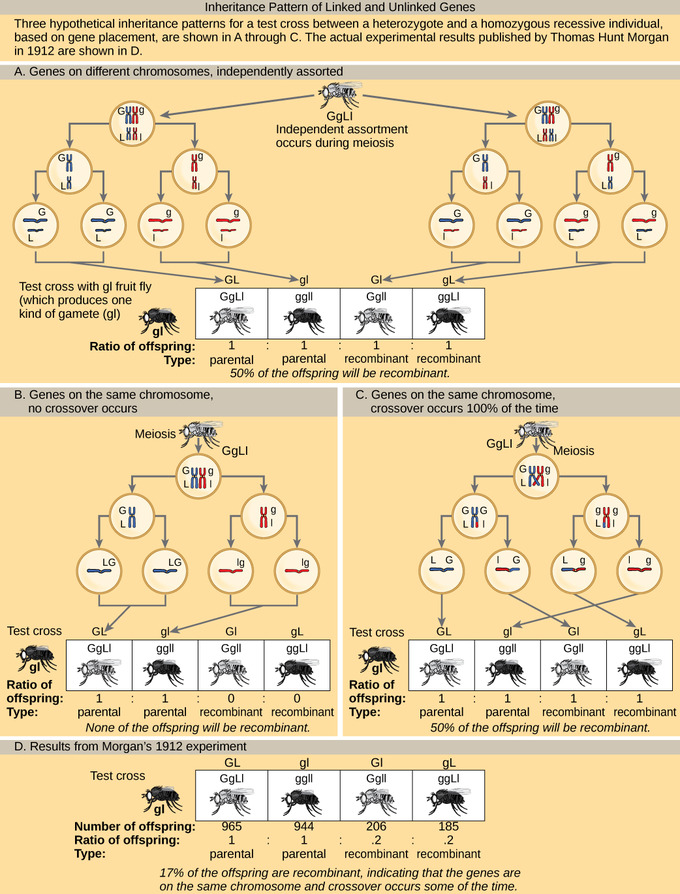
Inheritance Patterns of Unlinked and Linked Genes: In (a), two genes are located on different chromosomes so independent assortment occurs during meiosis. The offspring have an equal chance of being the parental type (inheriting the same combination of traits as the parents) or a nonparental type (inheriting a different combination of traits than the parents). In (b), two genes are very close together on the same chromosome so that no crossing over occurs between them. The genes are, therefore, always inherited together and all of the offspring are the parental type. In (c), two genes are far apart on the chromosome such that crossing over occurs during every meiotic event. The recombination frequency will be the same as if the genes were on separate chromosomes. (d) The actual recombination frequency of fruit fly wing length and body color that Thomas Morgan observed in 1912 was 17 percent. A crossover frequency between 0 percent and 50 percent indicates that the genes are on the same chromosome and crossover occurs some of the time.
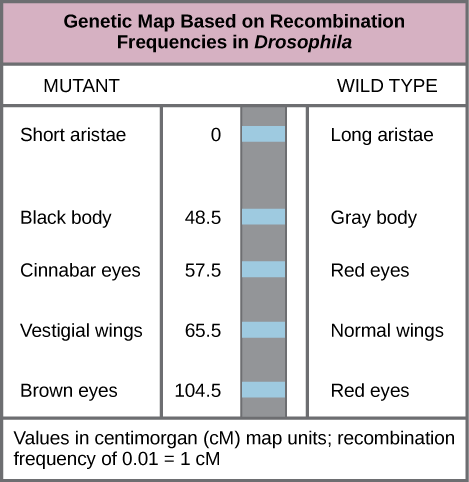
Genetic Maps: This genetic map orders Drosophila genes on the basis of recombination frequency.
By representing alleles in a linear map, Sturtevant suggested that genes can range from being perfectly linked (recombination frequency = 0) to being perfectly unlinked (recombination frequency = 0.5) when genes are on different chromosomes or genes are separated very far apart on the same chromosome. Perfectly unlinked genes correspond to the frequencies predicted by Mendel to assort independently in a dihybrid cross. A recombination frequency of 0.5 indicates that 50 percent of offspring are recombinants and the other 50 percent are parental types. That is, every type of allele combination is represented with equal frequency. This allowed Sturtevant to calculate distances between several genes on the same chromosome.
Chromosomal Structural Rearrangements
Structural rearrangements of chromosomes include both inversions and translocations, which may have detrimental effects on an organism.
Learning Objectives
Describe the various types of structural rearrangements of chromosomes and how they can impact an organism
Key Takeaways
Key Points
- A chromosome inversion is the detachment, 180° rotation, and reinsertion of part of a chromosome; this may have no effect on the organism, but if the inversion occurs within a gene or moves a gene away from its regulatory elements it can have an adverse effect.
- Pericentric inversions include the centromere, while paracentric inversions occur outside of the centromere; a pericentric inversion can change the length of the chromosome arms above and below the centromere.
- A pericentric inversion on chromsome 18 appears to have been involved in the evolution of humans.
- A translocation occurs when a segment of a chromosome dissociates and reattaches to a different, nonhomologous chromosome and can be benign or detrimental; in reciprocal translocations, there is no gain or loss of genetic information, so these are usually benign.
Key Terms
- inversion: a segment of DNA in the context of a chromosome that is reversed in orientation relative to a reference karyotype or genome
- translocation: a transfer of a chromosomal segment to a new position, especially on a nonhomologous chromosome
Chromosomal Structural Rearrangements
Cytologists have characterized numerous structural rearrangements in chromosomes, but chromosome inversions and translocations are the most common. Both are identified during meiosis by the adaptive pairing of rearranged chromosomes with their former homologs to maintain appropriate gene alignment. If the genes carried on two homologs are not oriented correctly, a recombination event could result in the loss of genes from one chromosome and the gain of genes on the other. This would produce aneuploid gametes.
Chromosome Inversions
A chromosome inversion is the detachment, 180° rotation, and reinsertion of part of a chromosome. Inversions may occur in nature as a result of mechanical shear, or from the action of transposable elements (special DNA sequences capable of facilitating the rearrangement of chromosome segments with the help of enzymes that cut and paste DNA sequences). Unless they disrupt a gene sequence, inversions only change the orientation of genes and are likely to have milder effects than aneuploid errors. However, altered gene orientation can result in functional changes because regulators of gene expression could be moved out of position with respect to their targets, causing aberrant levels of gene products.
An inversion can be pericentric and include the centromere, or paracentric and occur outside of the centromere. A pericentric inversion that is asymmetric about the centromere can change the relative lengths of the chromosome arms, making these inversions easily identifiable.
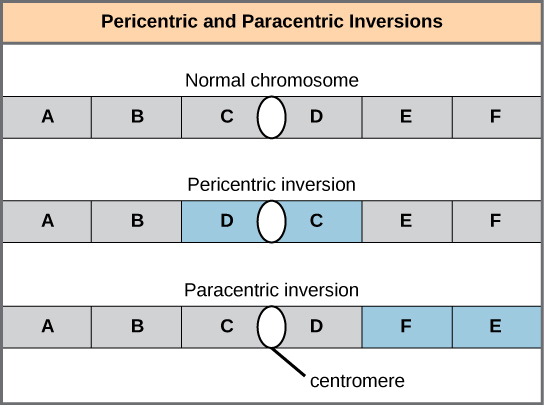
Inversions can be pericentric or paracentric: Pericentric inversions include the centromere, and paracentric inversions do not. A pericentric inversion can change the relative lengths of the chromosome arms; a paracentric inversion cannot.
When one homologous chromosome undergoes an inversion, but the other does not, the individual is described as an inversion heterozygote. To maintain point-for-point synapsis during meiosis, one homolog must form a loop, and the other homolog must mold around it. Although this topology can ensure that the genes are correctly aligned, it also forces the homologs to stretch and can be associated with regions of imprecise synapsis.
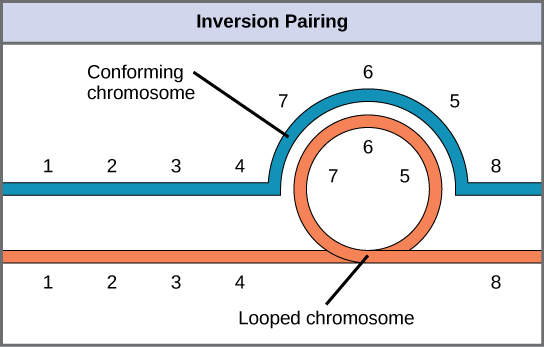
Inversion heterozygotes: When one chromosome undergoes an inversion, but the other does not, one chromosome must form an inverted loop to retain point-for-point interaction during synapsis. This inversion pairing is essential to maintaining gene alignment during meiosis and to allow for recombination.
Not all structural rearrangements of chromosomes produce nonviable, impaired, or infertile individuals. In rare instances, such a change can result in the evolution of a new species. In fact, a pericentric inversion in chromosome 18 appears to have contributed to the evolution of humans. This inversion is not present in our closest genetic relatives, the chimpanzees. Humans and chimpanzees differ cytogenetically by pericentric inversions on several chromosomes and by the fusion of two separate chromosomes in chimpanzees that correspond to chromosome two in humans.
The pericentric chromosome 18 inversion is believed to have occurred in early humans following their divergence from a common ancestor with chimpanzees approximately five million years ago. Researchers characterizing this inversion have suggested that approximately 19,000 nucleotide bases were duplicated on 18p, and the duplicated region inverted and reinserted on chromosome 18 of an ancestral human.
A comparison of human and chimpanzee genes in the region of this inversion indicates that two genes—ROCK1 and USP14—that are adjacent on chimpanzee chromosome 17 (which corresponds to human chromosome 18) are more distantly positioned on human chromosome 18. This suggests that one of the inversion breakpoints occurred between these two genes. Interestingly, humans and chimpanzees express USP14 at distinct levels in specific cell types, including cortical cells and fibroblasts. Perhaps the chromosome 18 inversion in an ancestral human repositioned specific genes and reset their expression levels in a useful way. Because both ROCK1 and USP14 encode cellular enzymes, a change in their expression could alter cellular function. It is not known how this inversion contributed to hominid evolution, but it appears to be a significant factor in the divergence of humans from other primates.
Translocations
A translocation occurs when a segment of a chromosome dissociates and reattaches to a different, nonhomologous chromosome. Translocations can be benign or have devastating effects depending on how the positions of genes are altered with respect to regulatory sequences. Notably, specific translocations have been associated with several cancers and with schizophrenia. Reciprocal translocations result from the exchange of chromosome segments between two nonhomologous chromosomes such that there is no gain or loss of genetic information.
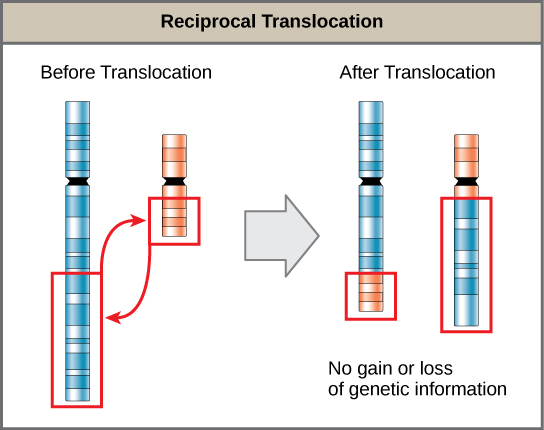
Reciprocal translocations do not involve loss of genetic information: A reciprocal translocation occurs when a segment of DNA is transferred from one chromosome to another, nonhomologous chromosome.
